Publications
For an updated list please visit Google Scholar. * denotes equal contribution.
2026
- TMLR 2026
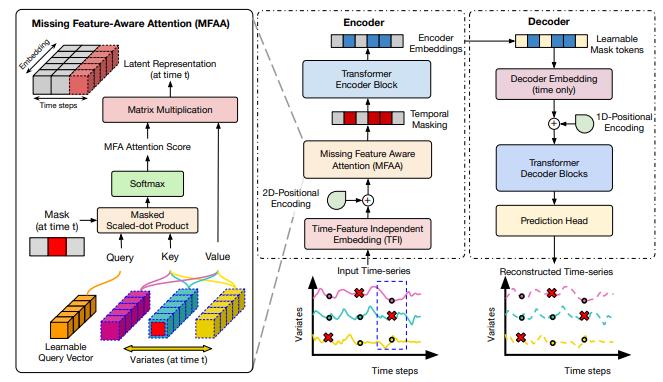 Investigating a Model-Agnostic and Imputation-Free Approach for Irregularly-Sampled Multivariate Time-Series ModelingTransactions on Machine Learning Research, 2026
Investigating a Model-Agnostic and Imputation-Free Approach for Irregularly-Sampled Multivariate Time-Series ModelingTransactions on Machine Learning Research, 2026
2025
- Under Review
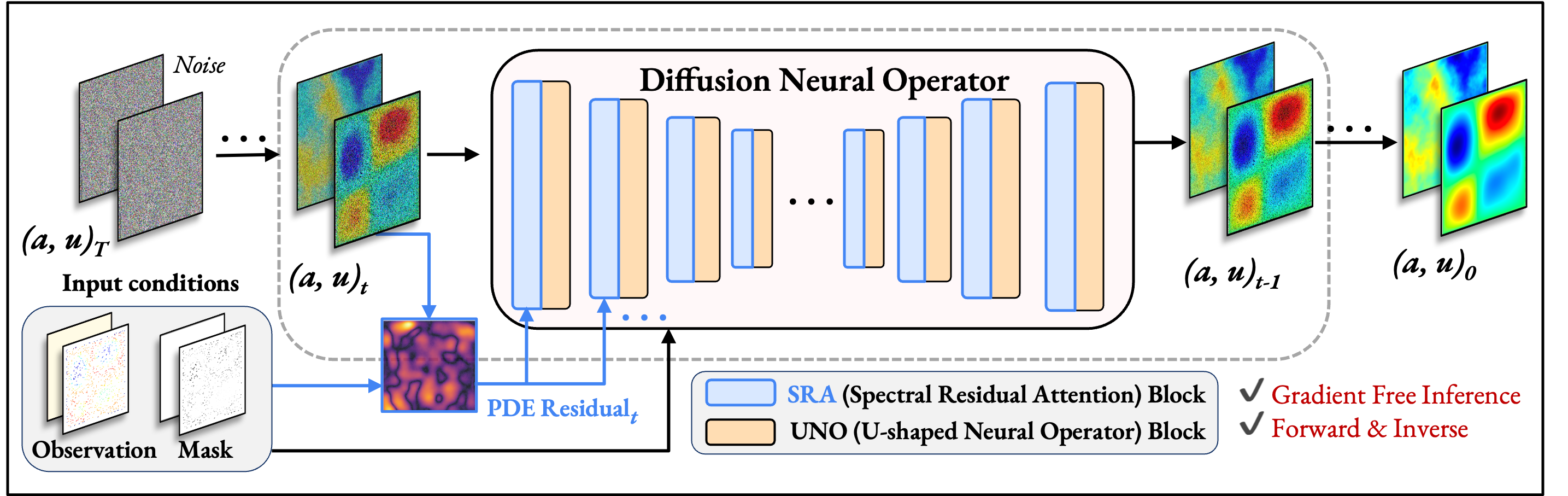 Beyond Loss Guidance: Using PDE Residuals as Spectral Attention in Diffusion Neural OperatorsUnder Review, 2025
Beyond Loss Guidance: Using PDE Residuals as Spectral Attention in Diffusion Neural OperatorsUnder Review, 2025 - CVPR W 2025
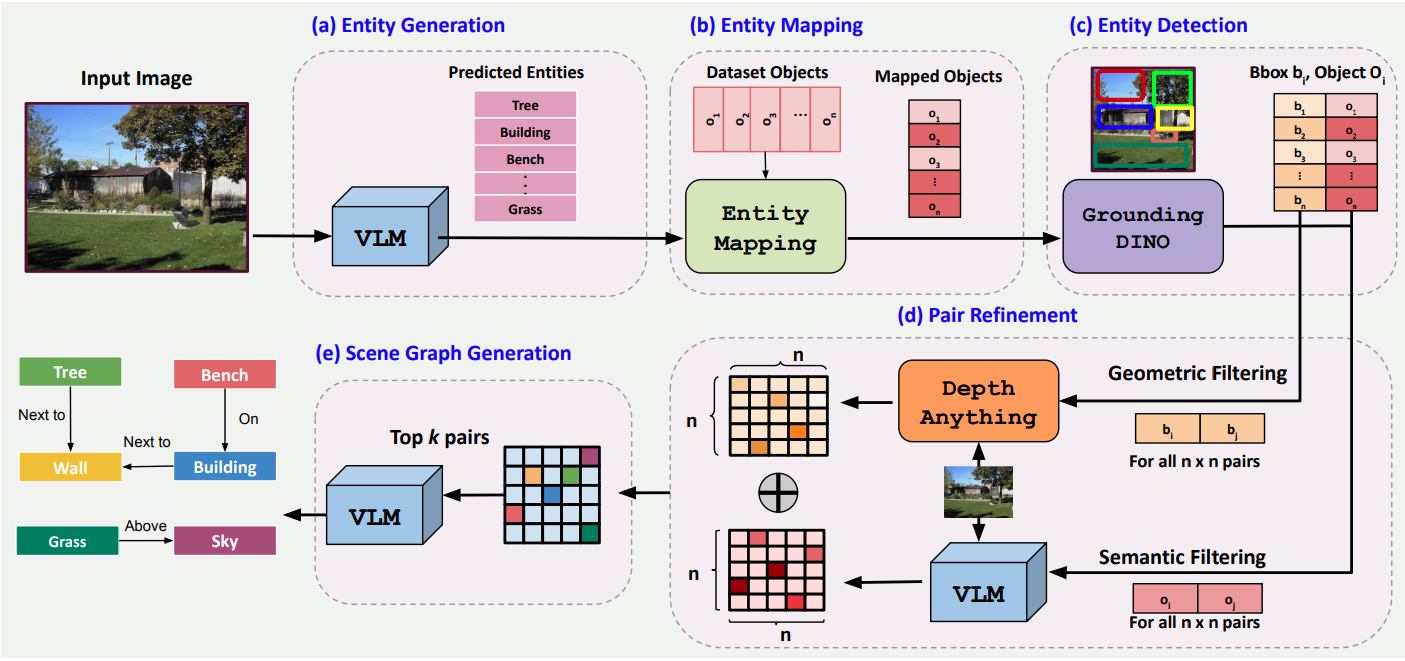 Open World Scene Graph Generation using Vision Language ModelsCV in the Wild Workshop, CVPR, 2025CVPR 2025 Workshop (CV in the Wild)
Open World Scene Graph Generation using Vision Language ModelsCV in the Wild Workshop, CVPR, 2025CVPR 2025 Workshop (CV in the Wild) - NeurIPS W 2025
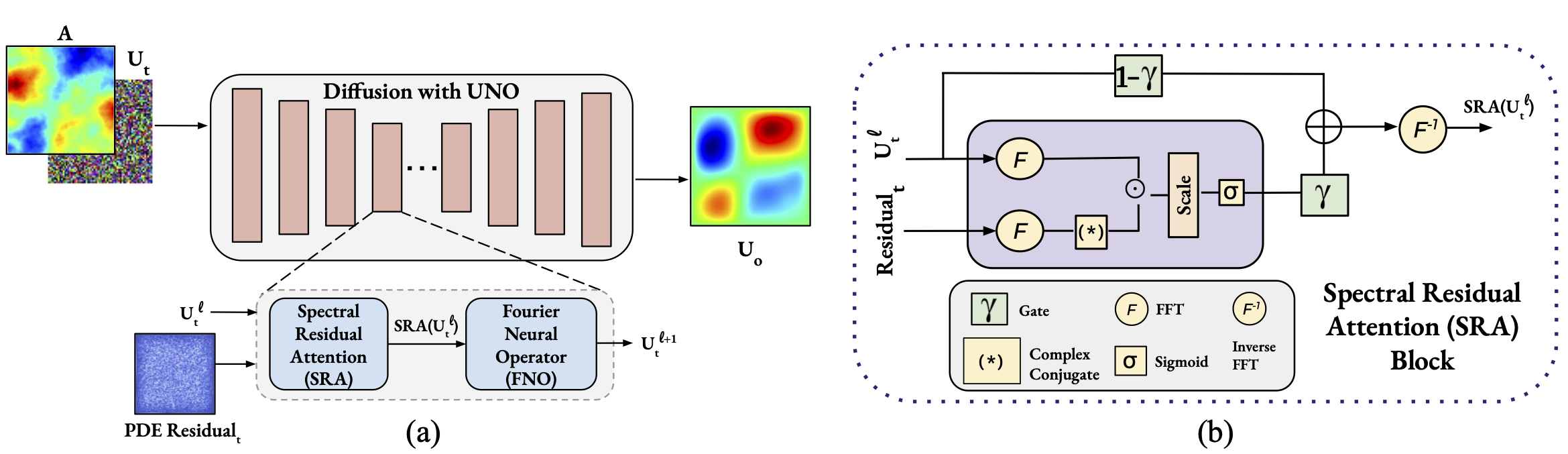 Investigating PDE Residual Attentions in Frequency Space for Diffusion Neural OperatorsML4Physics Workshop, NeurIPS, 2025Poster Presentation at ML4Physics Workshop
Investigating PDE Residual Attentions in Frequency Space for Diffusion Neural OperatorsML4Physics Workshop, NeurIPS, 2025Poster Presentation at ML4Physics Workshop - ICML W 2025
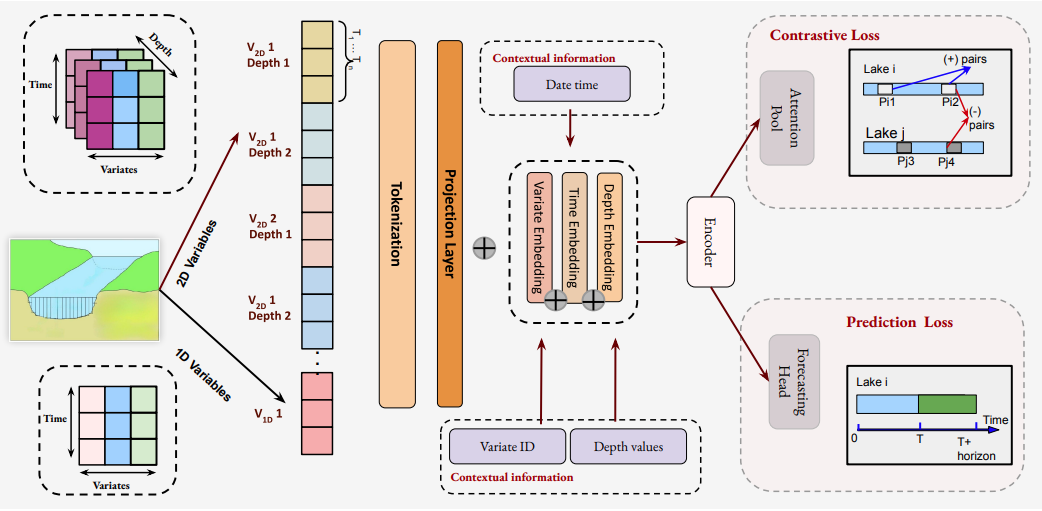 Toward Scientific Foundation Models for Aquatic EcosystemsUnder Review, 2025ICML 2025 Workshop (Foundation Models for Structured Data)
Toward Scientific Foundation Models for Aquatic EcosystemsUnder Review, 2025ICML 2025 Workshop (Foundation Models for Structured Data) - CVPR W 2025
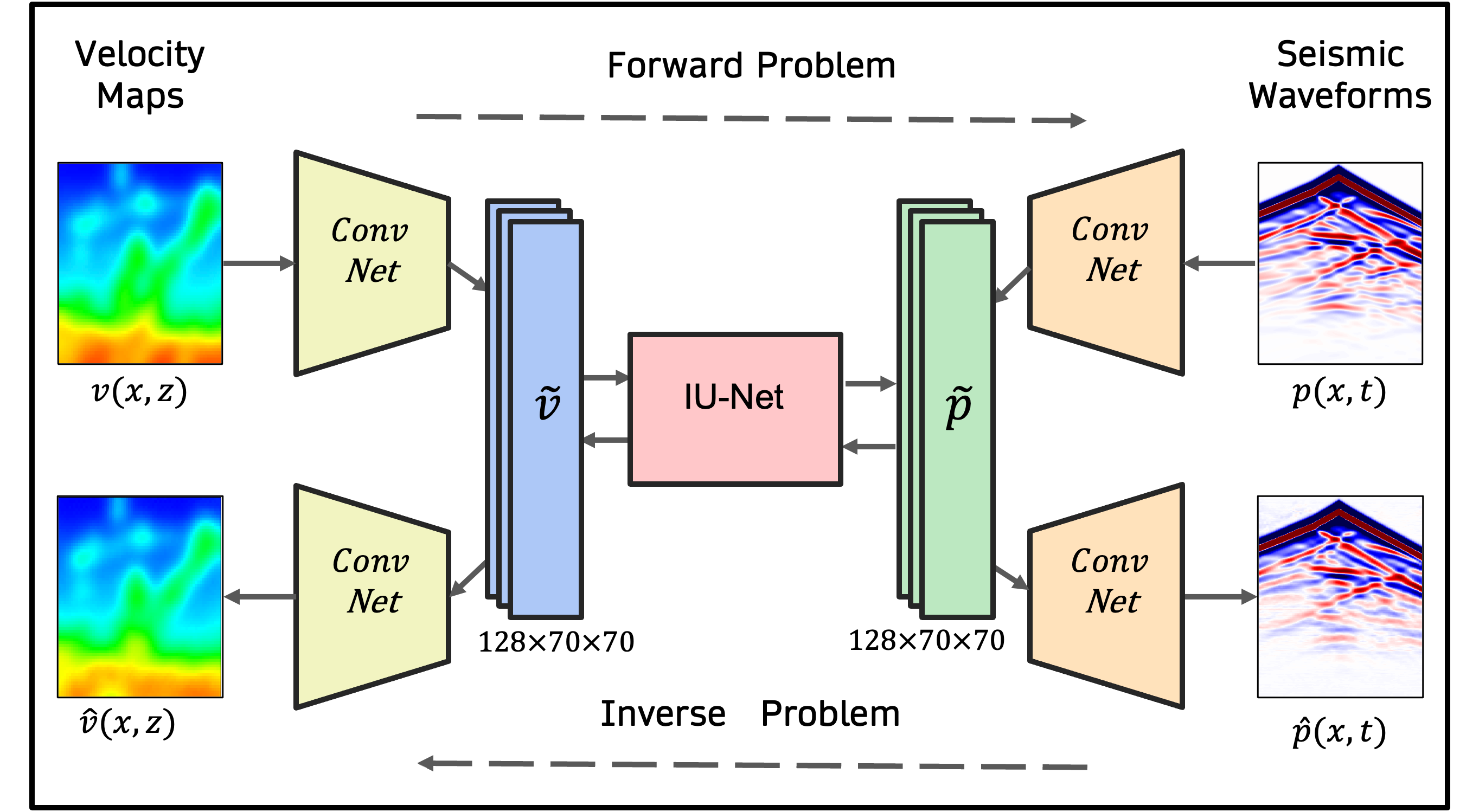
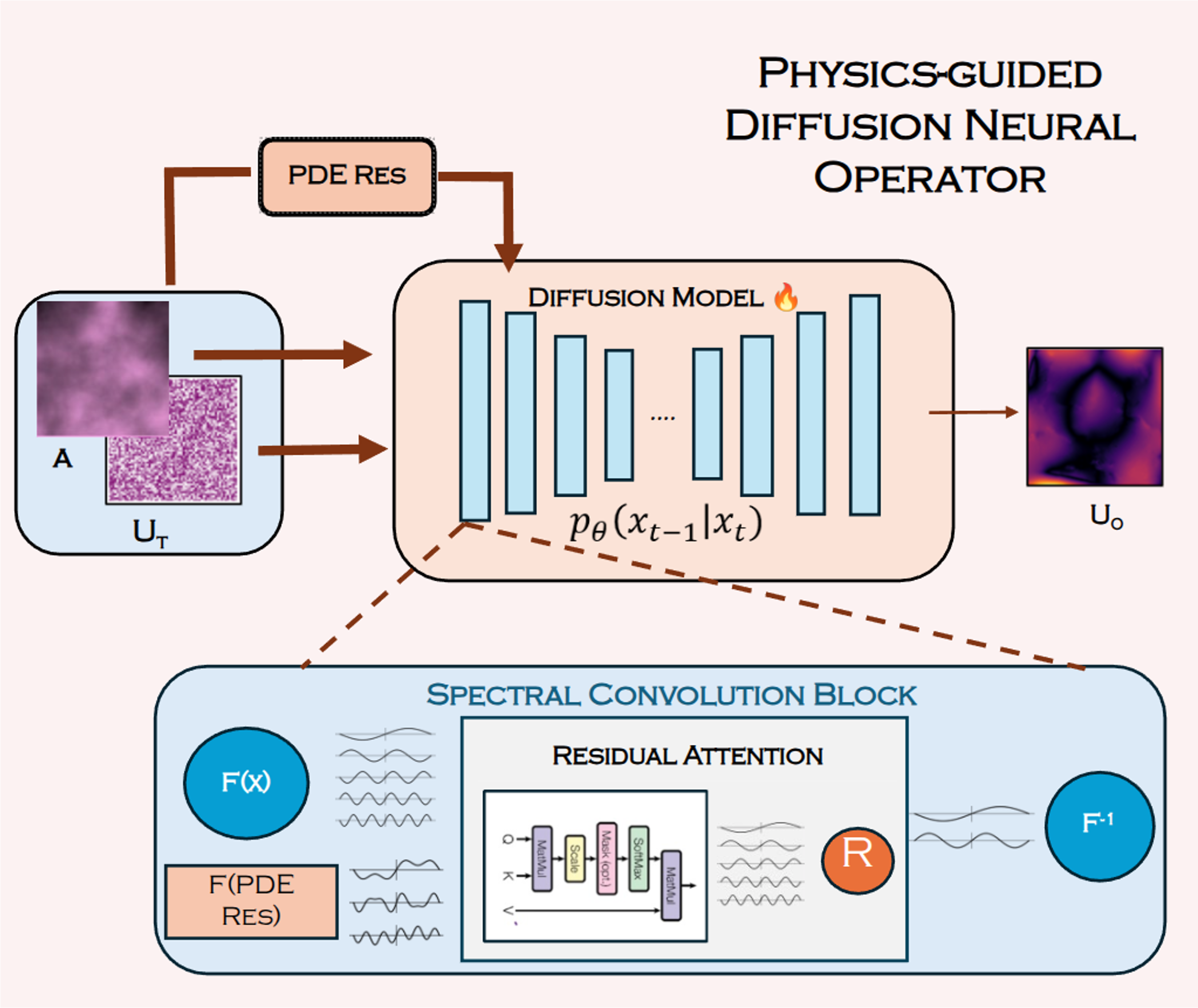 Physics-guided Diffusion Neural Operators for Solving Forward and Inverse PDEsCV4Science, CVPR, 2025Oral + Poster Presentation at CV4Science Workshop
Physics-guided Diffusion Neural Operators for Solving Forward and Inverse PDEsCV4Science, CVPR, 2025Oral + Poster Presentation at CV4Science Workshop - Under Review
 Investigating a Model-Agnostic and Imputation-Free Approach for Irregularly-Sampled Multivariate Time-Series ModelingUnder Review, 2025
Investigating a Model-Agnostic and Imputation-Free Approach for Irregularly-Sampled Multivariate Time-Series ModelingUnder Review, 2025